Abstract
The National Institutes of Health (NIH) are supporting the All of Us research program, a large multicenter initiative to accelerate precision medicine. The All of Us database contains information on greater than 400,000 individuals spanning thousands of medical conditions, drug exposure types, and laboratory test types. These data can be correlated with genomic information and with survey data on social and environmental factors which influence health. A core principle of the All of Us program is that participants should reflect the diversity present in the United States population.
The All of Us database has advanced many areas of medicine but is currently underutilized by primary care and public health researchers. In this Special Communication article, I seek to reduce the “barrier to entry” for primary care researchers to develop new projects within the All of Us Researcher Workbench. This Special Communication discusses (1) obtaining access to the database, (2) using the database securely and responsibly, (3) the key design concepts of the Researcher Workbench, and (4) details of data set extraction and analysis in the cloud computing environment. Fully documented, tutorial R statistical programming language and Python programs are provided alongside this article, which researchers may freely adapt under the open-source MIT license. The primary care research community should use the All of Us database to accelerate innovation in primary care research, make epidemiologic discoveries, promote community health, and further the infrastructure-building strategic priority of the family medicine 2024 to 2030 National Research Strategy.
- ADFM/NAPCRG Research Summitt 2023
- All of Us Database
- Cloud Computing
- Database
- Multicenter Studies
- Family Medicine
- Precision Medicine
- Primary Health Care
- Public Health
- Retrospective Studies
- Statistics
The All of Us Research Program and Primary Care Research
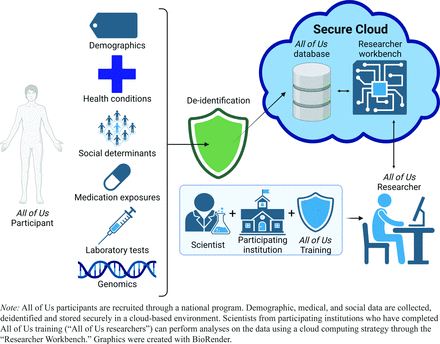
The All of Us research program is a large, multicenter initiative backed by the National Institutes of Health (NIH) to accelerate precision medicine.1 The All of Us research program has a target of recruiting 1 million or more participants to share data derived from electronic health records, biospecimens, and surveys in a centralized database. It also has as a core value that the set of participants “should reflect the broad diversity present in the United States population.”2 In its current form, it contains information for greater than 400,000 individuals spanning more than 25,000 distinct medical conditions, 29,000 drug exposure types, and 16,000 laboratory test types; 245,000 participants have small nucleotide polymorphism (SNP) and insertion/deletion (indel) variants annotated from short-read whole genomic sequencing, and an additional 1,000 participants have long-read whole genomic sequencing data available. All of Us survey data captures demographic and environmental factors variables that influence health, which allows these factors to be considered alongside biological variations to understand the development and trajectory of human disease more fully. (Figure 1)
Schematic diagram of the All of Us research platform.
The All of Us database has been used to make advances across broad areas of medicine including cancer3⇓⇓⇓⇓⇓⇓⇓⇓⇓–13; infectious disease (including COVID-19)14⇓⇓⇓⇓⇓⇓⇓⇓–23; pulmonary medicine10,24; cardiovascular medicine6,25⇓⇓⇓⇓⇓⇓⇓⇓–34; obesity35⇓–37; endocrine, diabetes and thyroid disorders31,34,38⇓⇓⇓⇓⇓–44; neurological, stroke, and neurocognitive disorders31,45⇓–47; dermatologic, breast and integumentary disorders6,8,16,21,24,42,45,48⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓–76; transplant medicine50; rheumatology, allergies and autoimmune medicine24,51,56,59,61,73,76,77; substance use disorders52,57,78,79; genomic medicine11,12,33,80; ophthalmology29,39,40,43,72,78,79,81⇓⇓⇓⇓–86; and gastroenterology and hepatology.13,17,56,87
Research using the All of Us database is in many cases directly relevant to the practice of primary care. Here I highlight 4 recent examples. First, recent work determined that the prevalence of hypertension in the All of Us cohort was 27.9%, and that this rate was similar to that previously observed in the National Health and Nutrition Examination Survey (NHANES) cohort.27 Epidemiologic studies of this nature are important to understand disease burden and influence policy about how resources should be allocated toward primary care. Second, “real-world” use of sodium glucose cotransporter 2 inhibitors (SGLT2i) and glucagon-like peptide 1 receptor agonists (GLP1-RA) were evaluated. Use of these medications was shown to be low, particularly among under-represented groups.41 In a third study, hypertension and type 2 diabetes were shown to be major risk factors for dementia.31 Multi-variable analysis also suggested that the interaction of race and ethnicity with hypertension indicates a need for targeted interventions in primary care settings in at-risk, structurally disadvantaged populations.31 Fourth, in the last study I highlight here, greater physical activity – as measured by step counts using Fitbit wearable devices – was associated with decreased risk of type 2 diabetes, and that multivariable analysis indicated that there was no significant modification of this effect by age, sex, body mass index, or sedentary time.44
I aim to highlight the promise of the NIH All of Us database for advancing primary care research. Concordant with the National Research Strategy that was recently developed at the 2023 National Family Medicine Research Summit,88 I encourage the primary care research community to use the All of Us database to enhance the practice of family and community medicine. To that end, I highlight detailed methods as a “tutorial walkthrough” to implementing an example project using the All of Us database. As an example, I explored the relationship between food insecurity and Vitamin B12 deficiency. The background, results, and clinical meaning of this association are discussed separately in a companion article. Here, I focus on replicability of the methods so that the barrier to use of the All of Us database is reduced for other primary care researchers.
The Researcher Workbench
All of Us makes data available to researchers through registration with the Researcher Workbench, available at https://www.researchallofus.org/data-tools/workbench/ (Figure 1). Researchers may gain access if they meet 2 conditions. First, they must complete training through the All of Us portal on issues related to data security and appropriate use of research data. A particular focus of this training is avoiding exacerbating pre-existing structural inequalities in health care. Completion of this training grants access to the “registered tier.” Additional training is required to access data within the “controlled tier,” which includes genomic data. Second, researchers must be members of an institution that has a data use and registration agreement with All of Us. This currently includes more than 600 participating institutions; an up-to-date list is available on their website at https://www.researchallofus.org/institutional-agreements/.
To create a project, researchers are required to publicly report the primary purpose of their project, which may be for research, educational, for-profit, or other purposes. Researchers must specify whether the project is: disease-focused research; methods development and validation; to develop controls, such as for comparisons with another data set from a different resource; social and behavioral research; population health or public health research; ethical, legal, and social implications research; or drug and therapeutics development research. The purpose of this project will be publicly displayed. Likewise, users must provide a publicly available summary of their research purpose by describing the scientific questions that are being evaluated by the study, and describing the relevance to science and public health. The scientific approach must also be briefly explained, and anticipated findings from the study must also be expressed. Researchers must report the population of interest to be studied, and if the population is a historically underrepresented population, this must be specified.
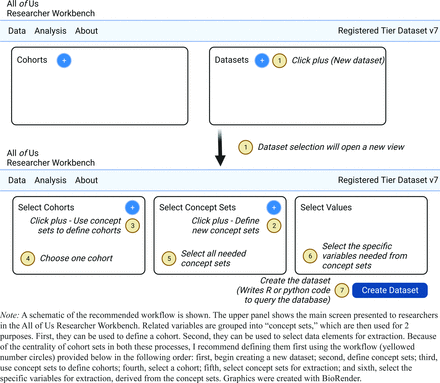
The Workbench uses “concepts,” “concept sets,” “values,” “cohorts,” and “datasets.” (Table 1). These are used to specify the population of interest and extract relevant variables from the database, as discussed in the next sections.
Glossary of Terms
Concept Sets
“Concept sets” can be defined to group related concepts together. This promotes ease of extraction of these variables when building datasets, and for ease of building cohorts (see next section). In this example, I define 3 concept sets: a vitamin B12 concept set, a food insecurity concept set, and a metformin-exposure concept set. The food insecurity concept set comprises 2 concept corresponding to whether the participant expressed (1) an experience of or (2) worry about food insecurity. The All of Us Researcher Workbench’s design implicitly suggests that users should first define a cohort, second define concept sets, and third define values for export. Because cohorts depend on data elements that are in concept sets, I instead suggest that users first define concept sets before defining a cohort (see Figure 2, which demonstrates the recommended workflow). Once a concept set is defined, it can then be readily used to define a cohort. Concept sets are therefore central to both cohort definition and dataset extraction.
Recommended workflow for defining concept sets, cohorts, and datasets.
Cohorts
Cohorts can be defined using a visual builder implementing Boolean logic (“AND” and “OR” relationships) between concepts and concept sets to define a population of interest. In my example about food insecurity and Vitamin B12 deficiency, I created a cohort of all participants who had both a Vitamin B12 measurement AND answered at least 1 of the 2 food insecurity questions on the social determinants of health survey. Specification of these constraints amounts to an AND condition between the Vitamin B12 concept set and the Food Insecurity concept set. For greater plausibility in the relationship between survey response and laboratory measurement, I wish to constrain these measurements to have occurred around the same time (eg, within a year). In principle, the visual interface in All of Us allows for expression of this sort of temporal constraint. In my hands, however, I observed that attempts to specify temporal queries often led to errors in the interface. I found it much easier to enforce temporal relationships between concepts during analysis in the cloud computing environment after exporting the dataset.
Dataset Extraction
Once concept sets and a cohort have been defined, a “dataset” can be constructed. A dataset is a group of concept sets applied to a cohort and requires choosing specific values to extract. The All of Us database contains a group of common concept sets that many projects may use, for example, demographics, and also allows users to define custom concept sets, as described above. For the Vitamin B12 and food insecurity example investigation, I used the above-discussed cohort and, from this, extracted (1) demographics, (2) vitamin B12 levels, (3) food insecurity-related answers to the social determinants survey, and (4) data on drug exposures (metformin). Note that concepts can be extracted even if they are not part of the cohort definition; in this example, presence/absence of metformin exposure was not part of the cohort definition because I want to capture the experience of individuals both with and without metformin exposure.
To extract the dataset All of Us writes a set of custom scripts in either R89 or Python90 (user selectable) containing a SQL database query to extract all specified values, and a few lines that save the results to the persistent cloud storage environment. These scripts can be run in the cloud computing environment, described below, as the first part of an analysis pipeline.
In this example, data are extracted from the database in 4 tables: person (demographics), measurements (Vitamin B12 levels), survey (answers to the food insecurity questions), and drug (metformin exposure). Concrete examples of integrating data from these tables is discussed in the accompanying example scripts available at https://github.com/djparente/AllOfUsExample and in Supplemental Materials.
Cloud Computing Environment
Data analysis is performed in a secure environment within the All of Us Researcher Workbench. Costs for using this environment are low, but not entirely free in the long-term, and depend on the computing resources required to conduct the project. The default environment recommended for general analysis by All of Us is a 4-CPU system with 15 GB of RAM, without a GPU. While the environment is running there is a cost of $0.20 per hour. This environment can be paused with a cost of “<$0.01” per hour. Persistent disk storage, at time of writing, cost $4.80 per month. Thus, for a project worked on for 80 computer hours over 6 months, a total computer cost of approximately $44.80 ($0.20/h x 80 hours + $4.80/mo x 6 months) would be incurred.
Use of fewer resources can reduce computer-time costs. For example, a 1-CPU system with 3.75 GB of RAM incurs an hourly cost of only $0.06 per hour. However, total costs are likely to be primarily driven by the duration storage is required (in this example, 6 months) rather than the costs of the computing environment. Using the least-resourceful system in the above example, the cost would be reduced only to $33.60. For my analyses on thousands of participants using descriptive techniques and logistic regression, I found the default configuration to be more than adequate. Greater computing resources are available for much higher costs per hour (eg, 96-CPU system with hundreds of GB of RAM and multiple graphical processing units [GPUs]). Conceivably such resource-intensive configurations might become important for very advanced applications, such as those driven by artificial intelligence and machine learning based on transformer architectures91 or local large language models;92⇓–94 or for advanced genomics research. More complex projects involving transferring exceptionally large volumes of database data (gigabytes or terabytes), or some workflows involving genomics, might incur additional costs. I suspect, however, that most primary care researchers will find the low-cost, default environment adequate for most of their purposes.
To defray initial start-up expenses, the All of Us researcher workbench provides $300 in free computing credits, which allows exploration of the research environment. Users can configure the computing environment to automatically suspend itself when idle for a duration (by default, 30 minutes) to prevent unintentional overuse of computing resources. This was completely adequate for my purposes; my initial projects familiarizing myself with the interface incurred <$30 in expenses, and I expect this experience to be true of most primary care researchers.
Data Preparation and Statistical Analysis
Within the workbench, researchers can use the R statistical computing language or the Python programming language to analyze data and perform statistical calculations. The default computing environments comes preloaded with many of the most used statistical and data operations packages for R and Python, but researchers are not limited to them. More packages can be installed, as needed, using the Python package index (PyPI) via pip, and through R’s “install.packages()” command. When a new computer environment is created, these packages may need to be reinstalled, so I recommend placing installation commands at the beginning of All of Us analysis scripts. The analysis environment is thus very flexible and can be readily adapted to many common workflows.
For the Vitamin B12 and food insecurity example, I have developed a set of example scripts that highlight data preparation in Python and statistical analysis in R. These example scripts are available at https://github.com/djparente/AllOfUsExample and in Supplemental Materials. These example applications demonstrate many common situations that researchers will likely encounter including, (1) transformation of survey data into usable formats, (2) extraction of numeric laboratory data, (3) extraction of drug exposure information, (4) enforcement of temporal constraints on the dataset (eg, considering only Vitamin B12 levels within 12 months of a survey response), (5) merging data together from various data tables (demographics, drug exposures, laboratory data, survey responses), (6) exclusion of outliers, (7) dichotomizing ordinal variables, and (8) conducting logistic regression and controlling for confounding variables. These example scripts may be freely adapted under the open-source MIT license for use in other All of Us projects. They also demonstrate how sophisticated analysis can be accomplished using only a small amount of code in the All of Us Researcher Workbench. In addition, because the example scripts use Python to do data preparation and R to do statistical analysis, it demonstrates how both frameworks can be used together and can exchange data through persistent disk cloud storage. If use of only 1 framework is desired, then data preparation and statistical analysis can be performed in either pure Python or pure R.
Users who are familiar with R and/or Python will find adapting to the cloud computing environment straightforward. Those familiar with SAS, SPSS or Stata will unfortunately not yet find these tools available in the All of Us cloud environment, but can find analogous statistical analysis functions in R. The addition of SAS, SPSS and Stata support to the All of Us environment in the future would further reduce barriers to use of the environment by researchers who might prefer these tools. Clinicians who have less experience with statistical analysis, R or Python may find collaboration with a data scientist or biostatistician to be mutually beneficial to all involved. Data scientists may benefit from clinician guidance toward questions likely to impact real-world clinical practice. Likewise, clinicians may benefit from data scientist/biostatistical expertise to ensure that analyses arrive only at robust, rigorous, and valid conclusions.
Human Subjects Protection
Collection of participant data are overseen by the NIH All of Us Institutional Review Board (IRB). The data portal has undertaken significant efforts to deidentify data and to reduce the risk of incidental reidentification. Data at the “Registered” access tier been (1) stripped of explicit identifiers and geolocation data, (2) undergone random shifting of dates, and (3) had removal of data from participants more than the age of 89 years. I expect that most analyses at the “Registered” access tier will be considered by local IRBs as “Not Human Subjects Research” as they are secondary data analyses on deidentified datasets. This example project was reviewed by the University of Kansas Medical Center IRB and determined to be Not Human Subjects Research. Nonhuman subjects research status may allow primary care researchers to analyze data rapidly with fewer regulatory barriers. By contrast, data in the “Controlled” access tier – including whole genomic sequencing, genotyping arrays, unshifted dates, and some suppressed demographic features – may more likely be considered “human subjects research” because those data elements are more likely to be considered by an IRB as “identifiable private information” under the 45 CFR 4695 definition of human subjects research. These considerations notwithstanding, I advise researchers to seek guidance from their local IRB on the protection of human subjects before initiating any All of Us-based projects.
Dissemination of Results
The All of Us research program requires researchers to notify the program at least 2 weeks before publications or presentations resulting from analyses of the data. Publications do not require approval from All of Us, but this notification is required to allow the program to prepare for media coverage or other issues that may be related to All of Us data. In addition, a final peer-reviewed manuscript must be deposited within PubMed Central immediately on acceptance for publication.
Primary Care Researchers Should Use All of Us to Accelerate Precision Medicine Research
In summary, the All of Us research program is an ambitious, multi-center effort to accelerate precision medicine. It has been used to advance many diverse areas within medicine.3⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓–87,96 Here, I highlight the potential for application of the All of Us database to address important issues within primary care, community medicine, and public health. I provide a detailed methodological template of using the database through the example of testing a hypothesis about Vitamin B12 deficiency and food insecurity. I hope this example and detailed instructions will reduce the technical “barrier to entry” to use of the All of Us platform, thereby driving greater primary care researcher engagement with it.
Leaders in family medicine recently met for the 2023 National Family Medicine Research Summit to create a national strategy for family medicine research between 2024 and 2030.88 This plan calls for research that is “whole-person, family, and community centered and improves health by enhancing health promotion, improving care for chronic diseases and advancing health care delivery, whereas including cross-cutting themes of health equity, technology, and team science.”88 Among the 3 strategic priorities identified by this plan is the development of a “national infrastructure for organizing and optimize family medicine research opportunities.”88 Within this strategic plan, primary care researcher use of the All of Us database can help accomplish some of the specific infrastructure-related objectives conceived of by the plan, viz.:
C2. Utilize a repository of clinical data to answer key questions in primary care, and
C5. Design and utilize distinctive methodology such as … big data analytics and machine learning (emphasis mine)
Primary care researchers will find that All of Us data can be used to improve the health of their patients and communities by uncovering associations between disease processes and social, environmental, and biological determinants of health. The All of Us database holds the potential to uncover new insights into disease processes that primary care clinicians frequently encounter, including obesity, type 2 diabetes, cardiovascular disease, cerebrovascular disease, and cancer. Primary care clinicians may also find benefit in research conducted by specialists on topics that are related to primary care. For example, there is a large and growing body of research works that use All of Us data to better understand dermatologic disease.6,8,16,21,24,42,45,48⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓–76 Primary care clinicians are often the “front line” clinicians who first encounter these disease processes. I nevertheless propose – and in concordance with the vision of the family medicine National Research Strategy88 – that primary care clinicians should not be content to merely observe specialist-conducted research on the All of Us database. Primary care involves implementing plans for whole persons, which is not adequately informed by the sum of research on individual organ systems. Primary care researchers must take a leadership role in using the All of Us database to enhance primary care and community health.
Many problems in primary care are strongly influenced by the social and environmental context of patients. Greater understanding of the associations between these factors and human disease will allow primary care clinicians to better care for whole persons. In doing so, they may also gain insight that will help alleviate pre-existing structural inequalities in health care that result from ongoing and historic biases. I invite the primary care research community to use All of Us to better understand these issues.
One future application for the All of Us database may be to accelerate research activities among family medicine resident physicians. Resident physicians must complete scholarly activities as part of ACGME requirements.97 I envision the construction of a “resident research pipeline” in which (1) resident physicians develop clinical questions inspired by their unique experiences during residency, (2) perform a literature search on what is already known about their questions, (3) construct a relevant cohort and data set in the All of Us portal, and (4) are guided in using bivariable and multivariable methods to answer their research question. Developing such a pipeline would allow residents to conduct potentially high-impact, multicenter research with results likely to generalize to other settings.
In closing, I call on the primary care research community to make use of the NIH All of Us database to accelerate innovation, make epidemiologic discoveries, and promote community health. Maximizing the generalizability and impact of primary care research conducted using the database may benefit from the formation of multi-institutional, national teams to develop and implement inquiries on topics of high clinical relevance to primary care. Moreover, I call on organizations dedicated to primary care – including the American Board of Family Medicine, the American Academy of Family Physicians, the Society of Teachers of Family Medicine, the Association of Departments of Family Medicine, and the North American Primary Care Research Group – to actively promote and devote resources to using the All of Us database to enhance the quality of primary care.
Notes
This article was externally peer reviewed.
This is the Ahead of Print version of the article.
Conflict of interest: None.
Funding: This work was funded by a contract from the American Academy of Family Physicians.
To see this article online, please go to: http://jabfm.org/content/00/00/000.full.
- Received for publication December 6, 2023.
- Revision received March 5, 2024.
- Revision received March 29, 2024.
- Accepted for publication April 1, 2024.
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.↵
- 79.↵
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵
- 88.↵
- 89.↵
- 90.↵
- 91.↵
- 92.↵
- 93.↵
- 94.↵
- 95.↵
- 96.↵
- 97.↵